- Search NCIBI Data
(e.g. diabetes, csf1r) - Login

NCIBI - "Try Our Tools" Virtual Workshops 2009 ARCHIVE
At our Annual Research Meeting in 2009, we ran tutorial workshops with over 75 participants. We have made these tutorials – 4 modules - and videos of the workshops available from this page below. Some tutorials are undergoing updating and will be updated by May, 2010.
Each tutorial shows how our tools facilitate and augment analysis for such questions as.
- What proteins interact and in what pathways?
- What compounds and reactions in a pathway may be associated with a set of genes?
- What interactions may play a mechanistic role in a disease?
- What articles are relevant to given genes, interactions or diseases?
- What genes are significantly enriched for a concept or for the same set of concepts?
Before you begin the workshops, you will need to download and install the application Cytoscape on your computer. To install Cytoscape please follow the instructions under "Pre-wokshop Cytoscape installation" below.
Virtual Workshops
Below are the list of files and the tutorial booklet you will need to download and access for each demonstration module during the workshop.
- Module 1: Finding Literature Based on Relevant Interactions and Pathways
- Module 2: Gene set enrichment testing and concept mapping
- Module 3: Visually exploring protein interaction networks and pathway associations
- Module 4: Exploring metabolomic data analysis tools
- Pre-workshop Cytoscape installation
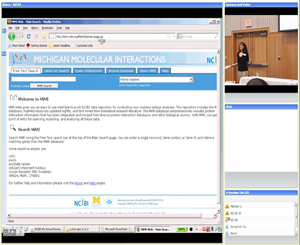 Module 1:
Module 1:
Finding Literature Based on Relevant Interactions and Pathways
(Featured tools: MiMI & MiMI Plugin for Cytoscape)
Related files:
Module1_RetrievingLit_Tutorial.pdf
Test_scenario_for_Module1.pdf
Launch Module 1 video
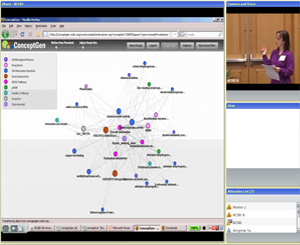 Module 2:
Module 2:
Gene set enrichment testing and concept mapping
(Featured tool: ConceptGen)
Related files:
Module2_ConceptGen_Tutorial.pdf
ConceptGen_Bipolar-Smoking_Genes.xls
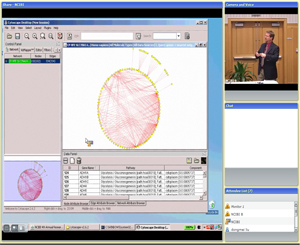 Module 3:
Module 3:
Visually exploring protein interaction networks and pathway associations
(Featured tool: MiMI Plugin for Cytoscape)
Related files:
Module3_MiMIPlugin_Tutorial.pdf
HemeANDSphingolipidGenes.txt
HemeOnlyGenes.txt
CytoiHemeAndSphingNoHdgReadInList.xls
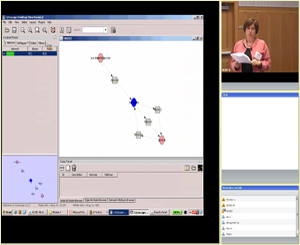 Module 4:
Module 4:
Exploring metabolomic data analysis tools
(Featured tool: MetScape Plugin for Cytoscape)
Related files:
Module4_ Metabolomics_Tutorial.pdf
5_compounds.txt
Metscape_36_compounds.txt
Metscape_prostate_exp_data.txt
Download all files in zip format
NCIBI_ARM_All_Workshop_Module_Files.zip (contains all workshop files listed above)
Pre-workshop Cytoscape installation
Cytoscape is an open source bioinformatics software platform for visualizing molecular interaction networks. Prior to the workshop please follow the below instructions to load Cytoscape on the computer you will be using for the workshop.
Cytoscape with workshop plugins requirements:
Any modern version of Mac OS X, Windows, or Linux
Minimum recommended hardware configuration: 1GHz processor, 512MB RAM
Java 5 SE or Java 6 SE Runtime Environment
- In general, Java SE 6 is faster than 5.
If your machine is compatible with the 6 series, please try version 6 here.
To launch the Cytoscape application:
Download cytoscape-ncibi-workshop.zip and unzip to your local computer.
Linux / MAC OS X:
Run cytoscape-runme.sh from the command line.
Windows XP or Vista:
Double-click cytoscape-runme.bat.
Alternatively:
You can pass the .jar file to Java directly using the command "java -Xmx512M -jar cytoscape.jar -p plugins". The -Xmx512M flag tells java to allocate more memory for Cytoscape and the -p plugins option tells cytoscape to load all of the plugins in the plugins directory. Loading the plugins is important because many key features like layouts, filters and the attribute browser are included with Cytoscape as plugins in the plugins directory.

 NCIBI on Facebook
NCIBI on Facebook NCIBI RSS Feed
NCIBI RSS Feed

