- Search NCIBI Data
(e.g. diabetes, csf1r) - Login

HighLights

Gibert S. Omenn, M.D., Ph.D.
DIRECTOR, of Center for Computational
Medicine and Bioinformatics
Professor of Internal Medicine,
Human Genetics, and Public Health

Arul Chinnaiyan
American Cancer Society Research
Professor
DIRECTOR, Michigan center for
Translational Pathology
Investigator, Howard Hughes Medical
Institute
Professor of Pathology and Urology
S.P. Hicks Endowed Professor
of Pathology

Matthias Kretzler, M.D.
Professor of Internal Medicine and
Research Professor,
Computational Medicine and
Bioinformatics, Medical School

Charles F. Burant, M.D., Ph.D.
Dr. Robert C. and Veronica Atkins
Professor of Metabolism
Associate Professor of Internal
Medicine Associate Professor of
Molecular and Integrative
Physiology

Melvin McInnis, M.D.
Thomas B. & Nancy Upjohn Woodworth
Professor of Bipolar Disorder and
Depression
Professor of Psychiatry
 Driving Biological Projects
Driving Biological Projects
Complex diseases are the next frontier in biomedical research. We are engaged in a fundamental transition from biomedical research focused on functions of single molecules or pathways to information-rich discovery science analyzing biological systems and their behavior on a genome-wide scale in health and disease. Successfully navigating that frontier requires new tools that aid researchers in locating, analyzing, applying, validating, and interpreting the massive amounts of data and text available to address their questions. Those tools are being developed now—and used now—in each of our Driving Biological Problems (DBP). These DBPs were selected based on compelling science, burden of disease, potential applications of our tools, and capabilities of the collaborating investigators.
The original four DBPs were the role of androgen signaling in prostate cancer progression; mechanisms and prevention of neuropathy, a major complication of type 1 diabetes; genomic heterogeneity of type 2 diabetes; and genomics of susceptibility to bipolar depression. After three years, the cancer biology DBP was transformed into studies of fusion genes; diabetic nephropathy succeeded neuropathy as the focus of type 1 and type 2 diabetic complications; drug abuse co-morbidities became the new focus for bipolar disorder; and metabolomics related to diabetes was added.
NCIBI's Suite of Tools and Data has been developed for the entire research community. Our DBP colleagues, as early-adopters, used NCIBI data integration, data mining, and pathways analyses to address major computational challenges in their research on fundamental biological interactions and clinical phenotypes. All of our current DBPs present important scientific and clinical challenges in common diseases.
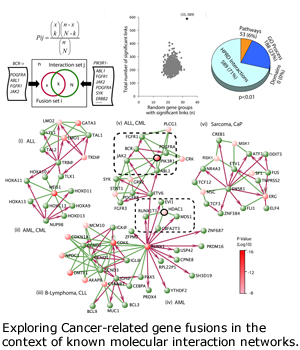
Gene Fusion in Cancers
Progression and metastasis of cancers cause most of the morbidity and mortality of cancers. DBP investigators from Arul Chinnaiyan’s group at the University of Michigan and from Leroy Hood’s group at the Institute for Systems Biology in Seattle productively investigated the many roles and pathways of androgen-receptor-mediated signaling in prostate cancers and prostate cancer cell lines. Based on bioinformatics analysis of outliers in gene expression datasets, the Chinnaiyan group discovered gene fusion events in prostate cancers. The most striking, involving the androgen-driven serine protease gene TMPRSS2 and the ETS family transcription factors ERG and ETV1, put the transcription and proliferation processes under androgen drive. These fusion genes are found in 70 percent of prostate cancers. Beginning in year four, we refocused our efforts in the prostate cancer DBP on fusion genes and on multi-level `omics analyses:
- Integrative informatics approaches have yielded additional novel fusion events in solid tumors (lung and prostate), which are then characterized with cell lines and clinical specimens. A general rule for these fusion-generating chromosomal rearrangements has been derived (Wang X et al, Nature Biotech, 2009).
- Complementary analyses linking transcriptomics, proteomics, and metabolomics through molecular concept maps have led to discovery of methyl-glycine (sarcosine) as both a biomarker and a potential key8 mediator of metastasis in prostate cancers (Sreekumar A et al, Nature 2009). We continue to develop our annotation capabilities for metabolomics with MetOntology and Metscape and it plug-in to Cytoscape.
- Comprehensive transcriptome analyses of CD133+ putative stem cells in human glioblastoma have utilized Oncomine, MiMI, and TALE, and are progressing to advanced sequencing of single cells.
Major Organ-Specific Complications of Diabetes
We have a concerted systems-level effort to identify genes, transcription factors, mechanisms, networks, and pathways which are causally involved in nephropathy (led by Matthias Kretzler) and neuropathy (Eva Feldman). We integrate molecular and physiological data to produce metabolic profiles of patient phenotypes (Charles Burant in the Michigan Metabolomics and Obesity Center). And we are extending the metabolomic studies, database, ontology, and visualization from cancers to diabetes (Alla Karnovsky, Chris Beecher). NCIBI data integration systems, data mining, and pathways analyses have transformed this work. We are:
- Identifying molecular signatures in diabetic nephropathy that would help define the active disease process in individual patients.
- Identifying shared transcriptional networks between human and murine diabetic nephropathy and neuropathy using advanced graph matching tools developed by cores 1 and 2, with striking findings for the JAK/STAT pathway.
- Disseminating use of our Nephromine database resource to the international renal research community.
- Exploring mechanisms of glucose-induced oxidative stress underlying neuropathy and nephropathy in diabetes mellitus and in experimental models for prevention. Patients with rapidly progressing disease in kidney and sural nerve biopsies, compared with those with slowly progressing disease, had activation of PPAR-gamma, EGFR, and FOS network nodes.
Co-Morbid Disease Associations of Bipolar Disorder
NCIBI is working with Melvin McInnis in the U-M Depression Research Center and national partners at Washington University (Laura Bierut) and the National Institutes of Health to develop an integrated translational research approach to the overlapping behavioral phenotypes of Bipolar Disorder and co-morbidities of Bipolar Disorder, especially involving substance abuse. These behavioral disorders are interrelated in complex ways. Collaboratively we are exploring integration at molecular, biochemical, and biological systems levels as well as disease phenotypes and endophenotypes. Major areas of focus include:
- Genetic factors of nicotine abuse and their relation to Bipolar Disorder and other behavioral and mood disorders
- Mining and curation of literature that would aid in prioritizing the genes in the chromosome 8q24 region for deep sequencing of that genomic region
- Analysis of gene expression findings in response to lithium treatment in relation to bipolar disorder and cocaine addiction, with in vitro studies using cultured human lymphoblastoid cells.

 NCIBI on Facebook
NCIBI on Facebook NCIBI RSS Feed
NCIBI RSS Feed

